GI Microbiome Panel
Understanding the state of the microbiome in the gut can help you manage your GI cases and provide better care.
Microbiome Testing – Overview
Canine or Feline
The microbiome has been an area of interest in humans and animals alike. Identifying the microbiota and its interaction with the host is key to understanding underlying causes to many of the chronic and persistent diseases we see in cats and dogs today. Now, it’s even easier to assess your patient’s microbiome with the GI Microbiome Panel.
| Test Methodology: | Next-Generation DNA Sequencing (NGS) |
| Species: | Canine or Feline |
| Sample Type: | Stool (fecal) – fresh *requires microbiome collection kit |
| Fasting: | Not Required |

What’s included?
- Complete bacteria and fungal quantification
- List of potential clinically relevant species
- Antibiotic resistance for relevant species, including a table of common antibiotics
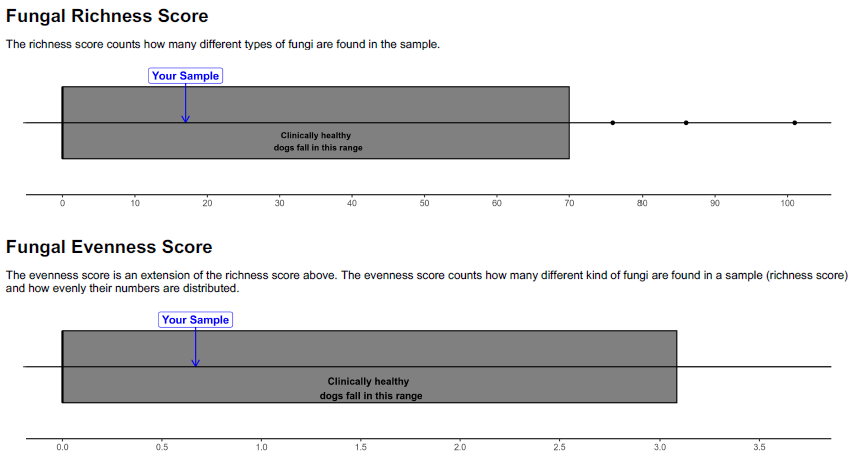
- Total Composition (Diversity)
- Richness Score
- Evenness Score
Which pets should test?
- ADR pets with undiagnosed GI signs
- Chronic GI distress
- Scratchy, Itchy, Red, or inflamed Skin
- Chronic Ear Infections/ Head shaking
- Excessive Licking
- Diarrhea or Vomiting with unknown source
Learn more about Microbiome Testing
Overview
The microbiome is vital to physiological homoeostasis in cats and dogs – influencing health and disease status. Beyond providing immunological assistance, the gut microbiome may also influence other physiological systems by competing for essential nutrients or digesting complex molecules to produce substrates for energy metabolism and cell signaling. Abnormalities in the GI microbiota can therefore also cause subtle but chronic effects, which can materialize into a range of metabolic and inflammatory diseases – especially chronic enteropathies.
Physiological Impact of Gut Microbiome
- Signaling the Immune System – pathogen resistance & immunological stimulation
- Prevent hypersensitization to environmental allergens
- Breakdown food components (e.g. fiber)
- Promote nutrient uptake and metabolism
- Synthesis of vitamins, bile acids, and amino acids
- Maintain the integrity of the gut lining
- Cell signaling – direct or indirect influence of biological systems
- Promote Angiogenesis
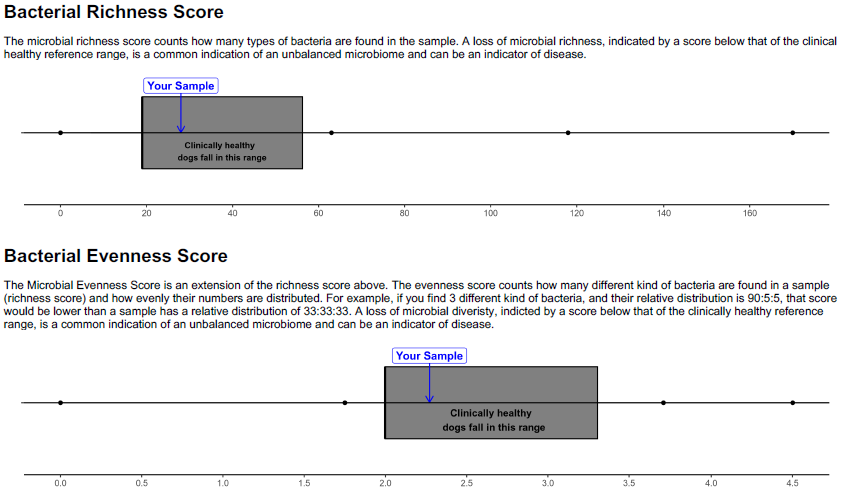
Microbiome Diversity
The composition of microbiome along the GI tract is influenced by age, diet, and many other environmental factors, however one of the defining features of a clinically healthy GI microbiome is diversity of species. Changes and imbalances in the microbiota, due to disease, lifestyle, or medications (e.g. antibiotics), affect its ability to perform the functions required.
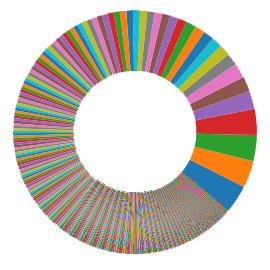
Clinically Healthy – Diverse Microbiota
Clinically Unhealthy – Loss of Diversity

Scientific research reveals that shifts in the specific bacterial and fungal species can contribute to diseases. Assessing the GI microbiota diversity as a clinical tool can assist by providing the necessary information for detecting susceptibility, providing diagnosis, and aiding the timely treatment of GI diseases.
The GI Microbiome Panel offers a complete look at the composition of the gut microbiota along with richness and evenness scores. When available, the relationship to a clinically normal reference group is provided.
NGS vs Cultures
Next Generation Sequencing (NGS) is a molecular based method that can sequence and detect the DNA of ALL microbes in a sample, without having to culture (grow) any of these organisms in a laboratory first. NGS is considered the gold standard for the detection and identification of microorganisms.
Traditional culture methods can only detect < 1% of all microorganisms, and most panels are targeted at certain pathogens- making educated guesses necessary. The NGS technology utilized by MiDog avoids these limitations, allowing for comprehensive detection of the bacteria and fungi, faster results, and no more “no growth” reports.
| GI Microbiome Panel* | Culture Testing | PCR Testing | |
|---|---|---|---|
| Diagnosis of ALL bacterial and fungal pathogens | YES | Only those that can grow in a lab | Limited to target panels |
| Antibiotic Resistance Profiling | YES | Only those that can grow in a lab | Limited to target panels |
| Pathogen Quantification | YES | Only those that can grow in a lab | Limited to target panels |
| Incubation/Growth Independent | YES | no | YES |
| Ambient Shipping/Storage Temp | YES | no | NO |
| Microbiome Profiling | YES | no | YES |
Importance of Antibiotic Resistances
The development and spread of antibiotic resistance results from antibiotic use, as well as intrinsic resistance exhibited by some pathogenic microorganisms. It is a growing concern for both humans and animals.
The GI Microbiome Panel can detect >46 acquired antibiotic resistance genes and reports intrinsic resistance of all microorganisms identified. By reporting both acquired and intrinsic antibiotic resistance, the panel helps guide veterinarians to choose the most appropriate antibiotic treatment, ultimately guiding good antibiotic stewardship.
Useful References
- https://www.mdpi.com/2079-6382/11/6/780
- https://www.sciencedirect.com/science/article/abs/pii/S1557506322000489
- https://www.sciencedirect.com/science/article/abs/pii/S1557506322000349?dgcid=coauthor
- https://avmajournals.avma.org/view/journals/ajvr/83/1/ajvr.21.02.0027.xml
- https://www.mdpi.com/2076-2615/11/12/3589#
- https://onlinelibrary.wiley.com/doi/abs/10.1111/vde.13017
- https://www.mdpi.com/2076-0817/10/7/904
- https://onlinelibrary.wiley.com/doi/full/10.1111/jvim.16104
- https://www.sciencedirect.com/science/article/pii/S0378113519314932
- https://avmajournals.avma.org/view/journals/ajvr/84/8/ajvr.23.03.0054.xml